準二項式迴歸¶
此筆記本演示如何使用自訂變異數函數和非二元資料,透過準二項式 GLM 族執行迴歸分析,以使用比例作為應變數。
此筆記本使用大麥葉枯病資料,該資料已在多本教科書中討論過。請參閱以下參考文獻之一
[1]:
import statsmodels.api as sm
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from io import StringIO
原始資料以百分比表示。我們將除以 100 以獲得比例。
[2]:
raw = StringIO(
"""0.05,0.00,1.25,2.50,5.50,1.00,5.00,5.00,17.50
0.00,0.05,1.25,0.50,1.00,5.00,0.10,10.00,25.00
0.00,0.05,2.50,0.01,6.00,5.00,5.00,5.00,42.50
0.10,0.30,16.60,3.00,1.10,5.00,5.00,5.00,50.00
0.25,0.75,2.50,2.50,2.50,5.00,50.00,25.00,37.50
0.05,0.30,2.50,0.01,8.00,5.00,10.00,75.00,95.00
0.50,3.00,0.00,25.00,16.50,10.00,50.00,50.00,62.50
1.30,7.50,20.00,55.00,29.50,5.00,25.00,75.00,95.00
1.50,1.00,37.50,5.00,20.00,50.00,50.00,75.00,95.00
1.50,12.70,26.25,40.00,43.50,75.00,75.00,75.00,95.00"""
)
迴歸模型是一個包含地點和品種效應的雙向加法模型。資料是一個完整的未重複設計,有 10 列(地點)和 9 欄(品種)。
[3]:
df = pd.read_csv(raw, header=None)
df = df.melt()
df["site"] = 1 + np.floor(df.index / 10).astype(int)
df["variety"] = 1 + (df.index % 10)
df = df.rename(columns={"value": "blotch"})
df = df.drop("variable", axis=1)
df["blotch"] /= 100
使用標準變異數函數擬合準二項式迴歸。
[4]:
model1 = sm.GLM.from_formula(
"blotch ~ 0 + C(variety) + C(site)", family=sm.families.Binomial(), data=df
)
result1 = model1.fit(scale="X2")
print(result1.summary())
Generalized Linear Model Regression Results
==============================================================================
Dep. Variable: blotch No. Observations: 90
Model: GLM Df Residuals: 72
Model Family: Binomial Df Model: 17
Link Function: Logit Scale: 0.088778
Method: IRLS Log-Likelihood: -20.791
Date: Thu, 03 Oct 2024 Deviance: 6.1260
Time: 15:58:09 Pearson chi2: 6.39
No. Iterations: 10 Pseudo R-squ. (CS): 0.3198
Covariance Type: nonrobust
==================================================================================
coef std err z P>|z| [0.025 0.975]
----------------------------------------------------------------------------------
C(variety)[1] -8.0546 1.422 -5.664 0.000 -10.842 -5.268
C(variety)[2] -7.9046 1.412 -5.599 0.000 -10.672 -5.138
C(variety)[3] -7.3652 1.384 -5.321 0.000 -10.078 -4.652
C(variety)[4] -7.0065 1.372 -5.109 0.000 -9.695 -4.318
C(variety)[5] -6.4399 1.357 -4.746 0.000 -9.100 -3.780
C(variety)[6] -5.6835 1.344 -4.230 0.000 -8.317 -3.050
C(variety)[7] -5.4841 1.341 -4.090 0.000 -8.112 -2.856
C(variety)[8] -4.7126 1.331 -3.539 0.000 -7.322 -2.103
C(variety)[9] -4.5546 1.330 -3.425 0.001 -7.161 -1.948
C(variety)[10] -3.8016 1.320 -2.881 0.004 -6.388 -1.215
C(site)[T.2] 1.6391 1.443 1.136 0.256 -1.190 4.468
C(site)[T.3] 3.3265 1.349 2.466 0.014 0.682 5.971
C(site)[T.4] 3.5822 1.344 2.664 0.008 0.947 6.217
C(site)[T.5] 3.5831 1.344 2.665 0.008 0.948 6.218
C(site)[T.6] 3.8933 1.340 2.905 0.004 1.266 6.520
C(site)[T.7] 4.7300 1.335 3.544 0.000 2.114 7.346
C(site)[T.8] 5.5227 1.335 4.138 0.000 2.907 8.139
C(site)[T.9] 6.7946 1.341 5.068 0.000 4.167 9.422
==================================================================================
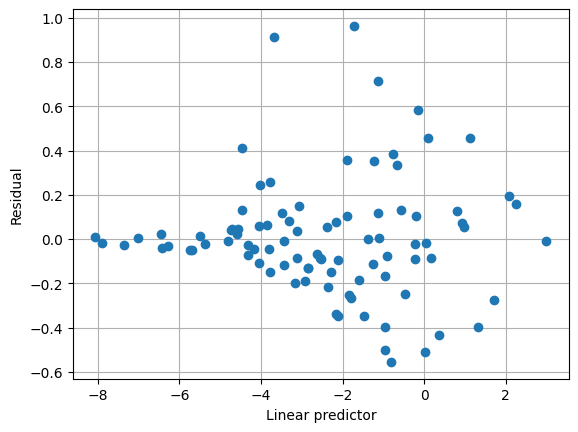
下面的圖顯示預設變異數函數沒有很好地捕捉變異數結構。另請注意,尺度參數估計值非常小。
[5]:
plt.clf()
plt.grid(True)
plt.plot(result1.predict(linear=True), result1.resid_pearson, "o")
plt.xlabel("Linear predictor")
plt.ylabel("Residual")
/opt/hostedtoolcache/Python/3.10.15/x64/lib/python3.10/site-packages/statsmodels/genmod/generalized_linear_model.py:985: FutureWarning: linear keyword is deprecated, use which="linear"
warnings.warn(msg, FutureWarning)
[5]:
Text(0, 0.5, 'Residual')
另一種變異數函數是 mu^2 * (1 - mu)^2。
[6]:
class vf(sm.families.varfuncs.VarianceFunction):
def __call__(self, mu):
return mu ** 2 * (1 - mu) ** 2
def deriv(self, mu):
return 2 * mu - 6 * mu ** 2 + 4 * mu ** 3
使用替代變異數函數擬合準二項式迴歸。
[7]:
bin = sm.families.Binomial()
bin.variance = vf()
model2 = sm.GLM.from_formula("blotch ~ 0 + C(variety) + C(site)", family=bin, data=df)
result2 = model2.fit(scale="X2")
print(result2.summary())
Generalized Linear Model Regression Results
==============================================================================
Dep. Variable: blotch No. Observations: 90
Model: GLM Df Residuals: 72
Model Family: Binomial Df Model: 17
Link Function: Logit Scale: 0.98855
Method: IRLS Log-Likelihood: -21.335
Date: Thu, 03 Oct 2024 Deviance: 7.2134
Time: 15:58:10 Pearson chi2: 71.2
No. Iterations: 25 Pseudo R-squ. (CS): 0.3115
Covariance Type: nonrobust
==================================================================================
coef std err z P>|z| [0.025 0.975]
----------------------------------------------------------------------------------
C(variety)[1] -7.9224 0.445 -17.817 0.000 -8.794 -7.051
C(variety)[2] -8.3897 0.445 -18.868 0.000 -9.261 -7.518
C(variety)[3] -7.8436 0.445 -17.640 0.000 -8.715 -6.972
C(variety)[4] -6.9683 0.445 -15.672 0.000 -7.840 -6.097
C(variety)[5] -6.5697 0.445 -14.775 0.000 -7.441 -5.698
C(variety)[6] -6.5938 0.445 -14.829 0.000 -7.465 -5.722
C(variety)[7] -5.5823 0.445 -12.555 0.000 -6.454 -4.711
C(variety)[8] -4.6598 0.445 -10.480 0.000 -5.531 -3.788
C(variety)[9] -4.7869 0.445 -10.766 0.000 -5.658 -3.915
C(variety)[10] -4.0351 0.445 -9.075 0.000 -4.907 -3.164
C(site)[T.2] 1.3831 0.445 3.111 0.002 0.512 2.255
C(site)[T.3] 3.8601 0.445 8.681 0.000 2.989 4.732
C(site)[T.4] 3.5570 0.445 8.000 0.000 2.686 4.428
C(site)[T.5] 4.1079 0.445 9.239 0.000 3.236 4.979
C(site)[T.6] 4.3054 0.445 9.683 0.000 3.434 5.177
C(site)[T.7] 4.9181 0.445 11.061 0.000 4.047 5.790
C(site)[T.8] 5.6949 0.445 12.808 0.000 4.823 6.566
C(site)[T.9] 7.0676 0.445 15.895 0.000 6.196 7.939
==================================================================================
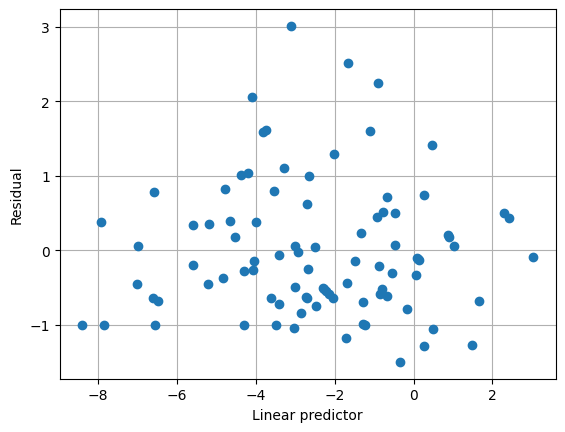
使用替代變異數函數,平均數/變異數關係似乎很好地捕捉了資料,並且估計的尺度參數接近 1。
[8]:
plt.clf()
plt.grid(True)
plt.plot(result2.predict(linear=True), result2.resid_pearson, "o")
plt.xlabel("Linear predictor")
plt.ylabel("Residual")
/opt/hostedtoolcache/Python/3.10.15/x64/lib/python3.10/site-packages/statsmodels/genmod/generalized_linear_model.py:985: FutureWarning: linear keyword is deprecated, use which="linear"
warnings.warn(msg, FutureWarning)
[8]:
Text(0, 0.5, 'Residual')
上次更新:2024 年 10 月 03 日